Mission: to support clinical investigators with state-of-the-art bioinformatics approaches for clinical research studies.
Powerful bioinformatics methods and tools now exist that can provide the infrastructure to standardize and organize complex data. Consequently, researchers can query across formerly disparate datasets to analyze and summarize data using new visualization techniques, removing many of the barriers for translational studies. Utilizing these methods and tools as well as drawing on experience using high dimensional data for research, the VERITY Bioinformatics Core faculty and staff will provide support and guidance to advance clinical research studies for rheumatic and musculoskeletal disease research. Moreover, the research questions arising from VERITY will present new methodologic challenges, driving advancement of bioinformatics methods and foster new collaborations.
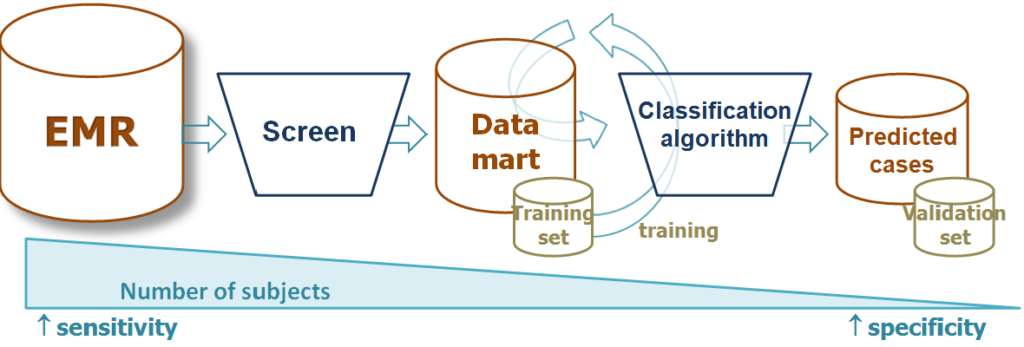
The VERITY Bioinformatics Core offers assistance with the following:
- Methods
- Developing phenotype algorithms using electronic health record data
- Note processing with NLP
- Create list of potential variables or features for the algorithm
- Development of phenotype algorithm via machine learning
- Algorithm validation
- Phenome-Wide Association Studies
- Standard PheWAS with ICD Codes
- Provide guidance on integrating ICD9 and 10 data for PheWAS
- MAP PheWAS with improved phenotype definitions incorporating NLP
- Study design and project planning using EMR data
- HLA imputation
- Developing phenotype algorithms using electronic health record data
- Extracting information from narrative data
- Extract information from narrative text notes using NLP
- Extract numerical data from notes, e.g. EF (using EXTEND, see below)
- Adapt methods or tools surrounding data extraction using NLP
- Tools
- Data for Clinical Research
- VERITY DATA Commons- bioinformatics platform for EHR based RA cohort integrated with BWH resources where users can search for summary data on EHR elements such as ICD codes, medications, NLP data on treatments, and availability biosamples at Partners Healthcare; click here to request access to the data.
- Consulting in areas related to the above Core services
*Please note – While the VERITY Bioinformatics Core assists investigators in applying algorithms and processing notes with NLP, the Core does not generally provide user support for on how to run the methods and tools. Most of the approaches require specific background training, and teaching investigators on how to use these tools and approaches is beyond the scope of the Core.
EHR-based Lung Cancer Progression Study – David Christiani, Massachusetts General Hospital
Phenotype pipeline development for the study of Ankylosing spondylitis (AS) – Steven Zhao, University of Liverpool
Algorithm development for classification of pseudogout using EHR data – Sara Tedeschi, Brigham and Women’s Hospital
Classification of autoinflammatory syndrome (AIS) using EHR data – Aleksander Lenert, University of Kentucky
Customization of CHANL for medical record review of patients with Multiple Sclerosis – Zongqi Xia, University of Pittsburgh
Linking Medicare data with Partners patients diagnosed with rheumatoid arthritis – Seoyoung Kim, Brigham and Women’s Hospital
Customizing CHANL to study CVD in patients with SLE – Karen Costenbader, Brigham and Women’s Hospital
Bioinformatics Resource Core Request Process

To access core services, you must be a VERITY member. More information about VERITY membership.
